There is a small disclaimer about this post. Obviously, this is a serious question which has left a lot of researchers befuddled, and conspiracy theories have been woven into an intricate web of deceit and lies. However, I must make it clear that I am not a laboratory scientist and I have based my opinion, summarized in this blog post, based on the available evidence and published papers. If there are any interesting, new evidence emerging in this area, which I have not taken cognizance of, do write in and let me know!
There has been a lot of speculation around the origin of the SARS-CoV-2 virus, and some have claimed that this was a laboratory generated virus, and not of natural origin. Recent phylogenomic and genetic evidence seem to indicate that the virus most likely evolved in nature and has been passed on from bats to humans through a yet to be identified intermediate host. Evidence and details on this issue have been discussed in depth in the previous blog post on the source of the new coronavirus.
If the current virus was generated in a lab, it is likely that one of the existing reverse-genetic systems available for betacoronaviruses would be used.[1] However, studying the genetic information of the SARS-CoV-2 isolates provides irrefutable proof that the virus is not derived from any existing virus backbone.[2]
Researchers have, instead, proposed that the virus underwent natural selection in either an animal host prior to zoonotic transmission to a human, or in a human after zoonotic transmission, to gain its virulent genetic footprint.[3] This assertion strongly indicates towards the fact that the virus was naturally circulating, and spilled over across the species barrier, and then underwent human to human transmission, eventually resulting in the current pandemic.
However, there remains a small possibility that the virus acquired the mutations in the receptor-binding domains (RBD), which gives it the unique genetic signature, during adaptation to passage in cell culture. While this is a remote and theoretical possibility, this has been previously documented in studies of SARS-CoV.[4] However, given that there have been viral isolates with SARS-CoV-like coronaviruses from pangolins which have shown to possess nearly identical RBDs, the biological plausibility that the virus originated in nature, within animals, is a stronger one, than the much discussed theory of a laboratory-escaped virus.
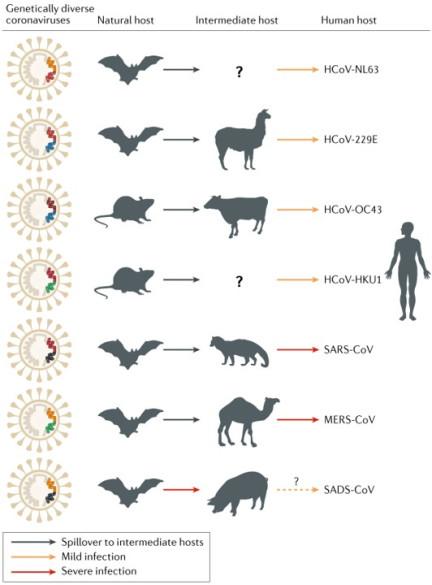
From Cui et al:
Severe acute respiratory syndrome coronavirus (SARS-CoV) is a new coronavirus that emerged through recombination of bat SARS-related coronaviruses (SARSr-CoVs) [5]. The recombined virus infected civets and humans and adapted to these hosts before causing the SARS epidemic [6,7]. Middle East respiratory syndrome coronavirus (MERS-CoV) likely spilled over from bats to dromedary camels at least 30 years ago [8] and since then has been prevalent in dromedary camels. HCoV-229E and HCoV-NL63 usually cause mild infections in immunocompetent humans. Progenitors of these viruses have recently been found in African bats [9,10], and the camelids are likely intermediate hosts of HCoV-229E [10,11]. HCoV-OC43 and HKU1, both of which are also mostly harmless in humans, likely originated in rodents. Recently, swine acute diarrhoea syndrome (SADS) emerged in piglets. This disease is caused by a novel strain of Rhinolophus bat coronavirus HKU2, named SADS coronavirus (SADS-CoV) [12]; there is no evidence of infection in humans. Solid arrows indicate confirmed data. Broken arrows indicate potential interspecies transmission. Black arrows indicate infection in the intermediate animals, yellow arrows indicate a mild infection in humans, and red arrows indicate a severe infection in humans or animals.
Summary
(a) There are conjectures which claim that the SARS-CoV-2 virus was a lab-generated virus, and not a naturally occurring one.
(b) These are likely to be unfounded theories, as the available evidence proves genetic similarity with coronaviruses previously isolated from bats.
(c) Studying the genetic make-up SARS-CoV-2 isolates provides irrefutable proof that it is not derived from any existing virus backbone.
(d) It is hypothesized that the virus underwent natural selection in either an animal host prior to zoonotic transmission to a human, or in a human after zoonotic transmission, to gain its virulent genetic footprint.
References
[1] Almazán F, Sola I, Zuñiga S, et al. Coronavirus reverse genetic systems: infectious clones and replicons. Virus Res. 2014;189:262-270. doi:10.1016/j.virusres.2014.05.026
[2] Cui J, Li F, Shi ZL. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol. 2019;17(3):181-192. doi:10.1038/s41579-018-0118-9
[3] Andersen KG, Rambaut A, Lipkin WI, Holmes EC, Garry RF. The proximal origin of SARS-CoV-2. Nat Med. 2020;26(4):450-452. doi:10.1038/s41591-020-0820-9
[4] Sheahan T, Rockx B, Donaldson E, Sims A, Pickles R, Corti D, Baric R. Mechanisms of zoonotic severe acute respiratory syndrome coronavirus host range expansion in human airway epithelium. Journal of virology. 2008 Mar 1;82(5):2274-85.
[5] Hu B, Zeng LP, Yang XL, Ge XY, Zhang W, Li B, Xie JZ, Shen XR, Zhang YZ, Wang N, Luo DS. Discovery of a rich gene pool of bat SARS-related coronaviruses provides new insights into the origin of SARS coronavirus. PLoS pathogens. 2017 Nov 30;13(11):e1006698.
[6] Song HD, Tu CC, Zhang GW, Wang SY, Zheng K, Lei LC, Chen QX, Gao YW, Zhou HQ, Xiang H, Zheng HJ. Cross-host evolution of severe acute respiratory syndrome coronavirus in palm civet and human. Proceedings of the National Academy of Sciences. 2005 Feb 15;102(7):2430-5.
[7] Chinese SARS Molecular Epidemiology Consortium. Molecular evolution of the SARS coronavirus during the course of the SARS epidemic in China. Science. 2004 Mar 12;303(5664):1666-9.
[8] Müller MA, Corman VM, Jores J, Meyer B, Younan M, Liljander A, Bosch BJ, Lattwein E, Hilali M, Musa BE, Bornstein S. MERS coronavirus neutralizing antibodies in camels, Eastern Africa, 1983-1997. Emerging infectious diseases. 2014 Dec;20(12):2093.
[9] Huynh J, Li S, Yount B, Smith A, Sturges L, Olsen JC, Nagel J, Johnson JB, Agnihothram S, Gates JE, Frieman MB. Evidence supporting a zoonotic origin of human coronavirus strain NL63. Journal of virology. 2012 Dec 1;86(23):12816-25.
[10] Tao Y, Shi M, Chommanard C, Queen K, Zhang J, Markotter W, Kuzmin IV, Holmes EC, Tong S. Surveillance of bat coronaviruses in Kenya identifies relatives of human coronaviruses NL63 and 229E and their recombination history. Journal of virology. 2017 Mar 1;91(5).
[11] Corman VM, Eckerle I, Memish ZA, Liljander AM, Dijkman R, Jonsdottir H, Ngeiywa KJ, Kamau E, Younan M, Al Masri M, Assiri A. Link of a ubiquitous human coronavirus to dromedary camels. Proceedings of the National Academy of Sciences. 2016 Aug 30;113(35):9864-9.
[12] Zhou P, Fan H, Lan T, Yang XL, Shi WF, Zhang W, Zhu Y, Zhang YW, Xie QM, Mani S, Zheng XS. Fatal swine acute diarrhoea syndrome caused by an HKU2-related coronavirus of bat origin. Nature. 2018 Apr;556(7700):255-8.
