In this second overview of research presented at SABCS, I thought I would focus on the excitement building with regard to options in triple negative breast cancer (that is tumors that don’t express the estrogen receptor, progesterone receptor or HER2).
In the past, triple negative disease has been defined in terms of what proteins are NOT expressed, but at this meeting, we heard from Dr Jennifer Pietenpol how TNBC is actually a heterogenous disease with 7 subtypes based on a large scale gene expression analysis of more than 3000 tumors from 21 worldwide data sets (not a trivial bioinformatics effort!). The major take home talk from her fabulous talk is that most of these subtypes have particular targets that may be feasible for targeting. As an example, about 11% of TNBCs actually express the Androgen Receptor (called the Luminal Androgen Receptor subtype), which is known to be a key growth regulator in prostate cancer, but until now has not received much attention in other tumors. The availability of AR antagonists such as bicalutamide means that now it is possible to target AR in breast cancers as well, although more detailed characterization of its function in breast cancer is still needed. As such, there will shortly be a trial available at Vanderbilt, using bicalutamide in combination with a PI3K inhibitor (because this subtype is highly enriched for PIK3Ca mutations – in TCGA dataset 50% of the LAR subtype had PIK3Ca mutations vs 3% in TNBCs as a whole). In case you are interested in reading more, the majority of Dr Pietenpol’s data was recently published in the Journal of Clinical Investigation (Lehmann et al, see references below).
Another interesting talk in the TNBC session on Tuesday was by Trey Westbrook, a young professor at Baylor College of Medicine. His research is focused on using genetic screens to uncover new targets. This functional approach is complementary to many of the large scale genomic screens such as the TCGA and ICGC that are being performed since targets that inhibit tumorigenesis that overlap with gene-level changes (amplification/mutation) are more likely to be true drivers vs passenger changes. In addition, targets that are validated functionally allow us to narrow down a large number of potential molecules for further study, since drug development and even basic research to understand mechanism is quite expensive.
So with that background, I will describe some of Dr Westbrook’s findings from his basic screen using a shRNA library transfected into normal mammary cells that have been immortalized with telomerase and SV40. The goal was to figure out which shRNAs induce transformation of these immortalized cells, which is tested using a standard anchorage-independent growth assay. A total of 42 new tumor suppressors were uncovered in this screen, but he only presented data regarding one of them, that is PTPN12. PTPN12 is a phosphatase that is mutated in about 5% of TNBCs but when analyzed further is found to be down-regulated by other (unknown) mechanisms in about 60% of TNBC tumors. Next the lab asked what does PTPN12 do? Phosphatases act in opposition to kinases that add phosphate groups to other proteins ie they remove these phosphates, which usually (but not always) turns off a protein.
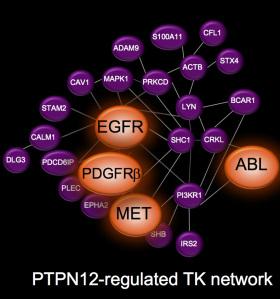
Figure 1: Network of proteins regulated by PTPN12
To figure out which proteins PTPN12 acts on, they did an elegant proteomic experiment using SILAC and compared the phospho-tyrosine proteome in cells with high PTPN12 levels and shRNA-depleted cells. What they found was that PTPN12 regulates a whole network of tyrosine kinase receptors including EGFR, HER2 and PDGFb, and that collectively these pathways drive tumorigenesis (see figure 1). This is exciting because there are currently available drugs against many of these pathways, so there is renewed enthusiasm in testing novel combinations of TKIs in this biomarker-defined subset (that is PTPN12 low). As a proof of principle preclinical study in xenograft tumors in mice, a combination of crizotinib and sunitinib to target cMet and PDGFRb (respectively) was tested. Individually each agent showed only very minimal tumor growth delay, however in combination tumor growth was halted, and the mice lived statistically longer (see key tumor growth rate in figure 2).
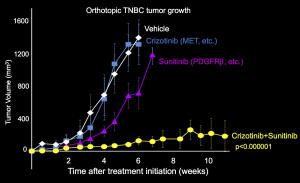
Figure 2: Tumor growth rate in mice treated with crizotinib and/or sunitinib
Once again, some of this work has been published recently in Cell, although this specific pharmacological combination was not presented in the publication (Sun T et al, see references)
References:
Lehmann B et al, “Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies” JCI 2011
Sun T et al, “Activation of multiple proto-oncogenic tyrosine kinases in breast cancer via loss of the PTPN12 phosphatase” Cell 2011
