[Guest post written by John Leavitt, Ph.D., Nerac, Inc., Tolland, CT.]
In 1985 my lab at the Linus Pauling Institute of Science and Medicine (LPISM) in Palo Alto, California started to work on an abundant protein of white blood cells (lymphocytes, macrophages, etc.) that mysteriously appeared in human tumor-derived cells of solid tissues (carcinomas, fibrosarcomas, melanomas, etc). I had noticed this phenomenon a few years earlier while at the National Institutes of Health. I also noticed that this protein appeared in oncogenic virus-transformed (SV40 virus) human fibroblasts, but the protein was not expressed in the normal fibrolasts.
I was intrigued by the fact that a major protein of circulating blood cells would be induced during solid tumor cell development because it is well known that solid tumor cells become more anchorage-independent and can circulate like white blood cells to metastasize to other organs. My colleague, David Goldstein, took the lead in examining the expression of this mysterious protein in different cell types of fractionated white blood cells. At the time this protein was assigned only a number (p219/p220) corresponding to its position in two-dimensional protein profiles. We found that this protein was abundantly expressed in all normal white blood cell types that we examined but it was not expressed in normal cells of solid tissues (Goldstein et al, 1985).
When David’s paper was submitted to Cancer Research, the reviews came back positive and the paper was accepted for publication, but one reviewer asked that we give the protein a name. I was thrilled by the thought of naming a protein and its gene which would immortalize our work, so I took on the serious task of coming up with a name that had lasting meaning. My theory was that this cancer marker contributed in some then-unknown way to the plasticity of the cytoplasm in solid tumor cells because of its normal presence in circulating white blood cells. Also, I had seen the great movie, The Graduate, with Dustin Hoffman and recalled that amusing scene depicted in the picture included below. So I named the protein “plastin” – the greatest new thing since sliced bread. :)

That same year, I met Steve Kent from Caltech at a meeting in Heidelberg, Germany. After hearing my talk, Steve suggested that we collaborate. He mentioned that a postdoctoral fellow in Leroy Hood’s lab, Dr. Ruedi Aebersold, was trying to develop a more sensitive protein sequencing method for purposes of determining snippets of amino acid sequences from small amounts of unknown proteins eluted from two-dimensional gels (protein profiles) like the gels that we used to characterize plastin in David’s paper. If we could get an accurate partial sequence of plastin, we could devise a nucleic acid probe based on the genetic code that could be used to clone a plastin “copy DNA” from a cDNA library. If the plastin cDNA was cloned, we could then define the protein and perhaps its function by determining the nucleic acid coding sequence in the clone.

Madhu Varma.
I gave Dr. Madhu Varma at LPISM the arduous task of isolating the plastin polypeptide “spot” for sequencing. Madhu cut out the stained spot from 140 two-dimensional gels, in effect purifying enough protein for sequencing by Ruedi at Caltech. Madhu succeeded and Ruedi produced eight short peptide sequences that could be used to develop short nucleic acid probes that would hybridize to the plastin cDNA clone isolated from a tumorigenic human fibroblast cDNA library.

Ching Lin.
Dr. Ching Lin at LPISM took one of the nucleic acid probes and immediately attempted to screen a tumorigenic fibroblast cDNA library. If we identified any clones that bound this probe, then we would perform a quick test to determine that we had cloned the plastin coding sequence. But science is full of surprises and we found that the first clone he isolated detected a gene product that was not in lymphocytes but only in normal human fibroblasts – in other words, it failed the test. This is where Ching’s brilliance took over. He was convinced that this first clone he had isolated was indeed a plastin coding sequence so he used this clonal DNA as a new probe against the tumorigenic fibroblast cDNA library. He isolated a new clone that passed the test and detected a gene that was expressed in lymphocytes and tumorigenic fibroblasts but not in normal human fibroblasts.
We performed other experiments that proved that we had cloned two different isoforms of plastin: L-plastin, expressed in lymphocytes and solid tumor-derived cells, and T-plastin that was expressed in normal solid tissues and co-expressed with L-plastin in tumor cells from solid tissues (Lin et al, 1988; Lin et al, 1990). Ultimately this work led to the complete characterization of the human plastin multigene family and verification that both isoforms were aberrantly expressed in various types of human tumors.
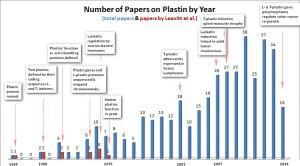
The figure at the top of this post maps the progression of discovery that followed our research, which began at the Pauling Institute in 1985. Our publications are shown in red in the graph and research published by other labs is shown in the blue bars.
Here are several plastin milestones discovered by other researchers:
- T-plastin is abundantly induced in Sezary lymphomas, a lethal T-lymphocyte cancer (Su et al, 2003);
- L-plastin induction in solid tumors contributes to invasive cancer growth and metastasis (Klemke et al, 2007);
- Mutations in T-plastin play a role in the genetic disease Spinal Muscular Atrophy (Oprea et al, 2008); and
- Most recently mutations in both L- and T-plastin promote re-growth of colon carcinomas following surgical resection of these tumors and chemotherapy (Ning et al, 2014).
These developments are more or less typical of the way science works. Progress in understanding complex phenomena like human cancer is the work of many scientists that builds on the observations of other scientists. This is just one example of the productive contributions in biomedical research that came about through early discovery research at LPISM in the 1980s.
