5-hydroxy-methyl CpGs (5-hmCs) were first discovered in 2009 and shown to be enriched in the brain, but remain a mysterious epigenetic mark, despite intriguing functional findings such as: environmental enrichment’s reduction of it, MeCP2′s preference for 5mc over 5hmc, and it’s possible role as an intermediate in demethylation. This new technique will aid their characterization by allowing absolute quantification and base-resolution localization of the marks. The technique also serves as a reminder of why you should pay attention in orgo, or at least why you should collaborate with people who did!
Now, Emory’s Peng Jin, has collaborated with University of Chicago chemist He Chuan to develop a new derivative of bisulfite sequencing, Tet-Assisted Bisulfite Sequencing (TAB-Seq) that distinguishes 5-hmcs from 5-mcs, as they describe in cell.
Yu (2012) Cell
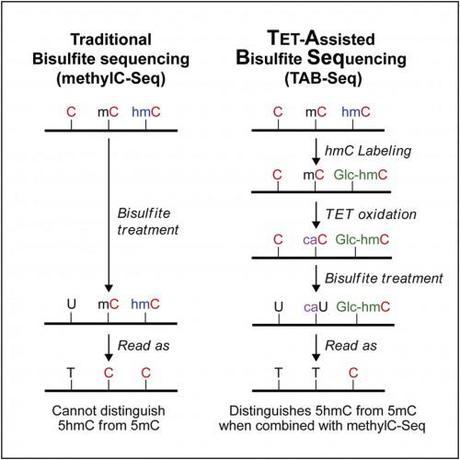
Traditional bisulfite sequencing (MethylC-Seq):
- Sequence the sample
- Treat the sample with bisulfite, which converts all non-methylated cytosines to uracils, but leaves 5-mCs and 5-hmCs as cytosines. (The even rarer base 5-carboxy-C (5-caC), is converted by bisulfite into 5-caU.)
- Resequence.
- Compare your first sequence to your second. You know the unmethylated cytosines from the first sequence will show up as Ts in the second sequence (because when they are amplified, they will be amplified as Thymidine not Uracil). The methylated and hydroxy-methylated cytosines which show up as Cytosines in both sequences.
TAB-seq
So, how can we differentiate 5-hmCs from 5-mCs? In a process that may remind you of all those organic chemistry synthesis problems, TAB-seq involves an extra step to protect the hydroxy-methylated cytosines from TET oxidation.
- Glucosylate 5hmC using β-glucosyltransferase (βGT).
- 5mC is oxidized to 5caC by with an excess of recombinant Tet1. (The blocked 5hmCs (β-glucosyl-5-hydroxymethylcytosine (5gmC) ) are not oxidized.)
- Treat the sample with bisulfite. This converts the Cs and 5-caCs to Us, but doesn’t effect the 5gmC.
- Resequence
- Compare back with traditional bisulfite sequencing. The 5-hmCs are the bases that show up as Cs in these two sequences. (5-mCs will show up as Ts in TAB-seq, but Cs in traditional. Unmodified Cs and CaCs will show up as Ts in both sequences.)
Validation and Findings
Validation of new techniques (proof that they work), is always important, and the paper shows that it works using mass spectrometry.
They validated it’s practicality, by using the technique to map 5-hmCs in human embryonic stem cells (hESCs) and mouse embryonic stem cells (mESCs). In hESCs they found 691,414 5hmCs with a false discovery rate of 5%. Interestingly, though mice have similarly sized genomes, they found much higher levels of 5hmCs–2,057,636–which they hypothesize is also due to the higher levels of Tet1 and Tet2 proteins.
So where are 5hmCs enriched, now that we can identify them precisely? H1 distal-regulatory elements including p300-binding sites (observed/expected [o/e] = 7.6), predicted enhancers (o/e = 7.8), CTCF-binding sites (o/e = 5.1)–CTCF is a transcriptional repressor that blocks interactions between promoters and enhancers and also plays a role in stopping the spread of heterochromatin, and DNase I hypersensitive sites (o/e = 3.4) which are associated with active gene expression. Because, 5-hmCs are enriched at enhancers, the authors speculate that 5-hmC may be specifically recognized by transcription factors as a core base in binding motifs.
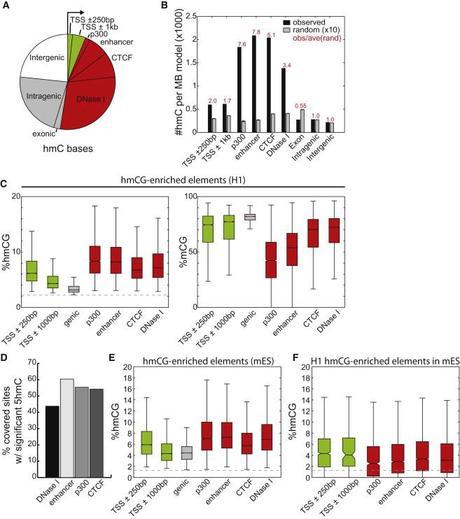
Genomic Distribution of 5hmC Sites. Yu (2012) Cell.
Many genes had significant enrichment of 5hmC, but lowly expressed genes had more than highly expressed. 5hmCs showed asymmetry, with more hydroxylation on strands where the CpG was surrounded by Gs. (A similar pattern wasn’t observed for 5mCs.)
5hmCs also tended to be enriched near low CpG areas
Previous findings that identified 5hmCs in high CpG areas, such as CpG island-containing promoters, but these findings are likely do to the bias of mapping techniques which can amplify frequent weak signals and overshadow sparse but strong ones. The present study, found that 5hmCs tended to be enriched in lower CpG areas, especially those with H3K4me3 or bivalent (H3K4Me3 and H3K27ac) chromatin modifications, but how 5hmC interacts with the histone code is still up in the air.
Conclusion
It will be interesting to see if the findings from, generalize to different cell-types, but since hESCs and mESCs showed similar patterns, it suggests that the regulation at least in stem cells is evolutionarily conserved.
It seems this tree will have bountiful fruit, weighing down the branches for some time. I’ll leave a final summary in the authors’ own words:
“We have developed a genome-wide approach to determine 5hmC distribution at base resolution and have generated base-resolution maps of 5hmC in both hESCs and mESCs. These maps provide a template for further understanding the biological roles of 5hmC in stem cells as well as gene regulation in general. In conjunction with methylC-Seq, the TAB-Seq method described here represents a general approach to measure the absolute abundance of 5mC and 5hmC at specific sites or genome-wide, which could be widely applied to various cell types and tissues.”
References
Kriaucionis, S., & Heintz, N. (2009). The nuclear DNA base , 5-hydroxymethylcytosine is present in brain and enriched in Purkinje neurons. Science, 324(5929), 929-930. (Free full text.)
Szulwach, K. E., Li, X., Li, Y., Song, C.-X., Wu, H., Dai, Q., Irier, H., et al. (2011). 5-hmC-mediated epigenetic dynamics during postnatal neurodevelopment and aging. Nature neuroscience, 14(12), 1607-16. Nature Publishing Group. doi:10.1038/nn.2959
Yu M, Hon GC, Szulwach KE, Song CX, Zhang L, Kim A, Li X, Dai Q, Shen Y, Park B, Min JH, Jin P, Ren B, & He C (2012). Base-resolution analysis of 5-hydroxymethylcytosine in the Mammalian genome. Cell, 149 (6), 1368-80 PMID: 22608086
Guo, J. U., Su, Y., Zhong, C., Ming, G.-li, & Song, H. (2011). Emerging roles of TET proteins and 5-hydroxymethylcytosines in active DNA demethylation and beyond. Cell Cycle, 10(16), 2662-2668. doi:10.4161/cc.10.16.17093
———-
You may also be interested in the brief article I wrote previously about 5-hmCs and a paper that showed that they are highly enriched in the cerebellum and hippocampus (10x higher than in stem cells), and they increase with age. Further, the authors showed that MeCP2–which strongly binds the unhydroxylated and more ubiquitously expressed version, 5-methyl CpGs–does not bind 5-hmCs. Overexpression of MeCP2 even seems to block TETs from converting 5-mCs into 5-hmCs.


