
Linus Pauling and Robert Corey examining models of protein structure molecules, ca. 1951.
[Part 1 of 3]
An article with the somewhat cumbersome title “The Structure of Proteins: Two Hydrogen-Bonded Helical Configurations of the Polypeptide Chain” appeared in the April-May, 1951 edition of the Proceedings of the National Academy of Sciences. The article was written by Linus Pauling, Robert B. Corey and Herman R. Branson, working collaboratively at the Gates and Crellin Laboratories of Chemistry at the California Institute of Technology and communicated to PNAS on Pauling’s fiftieth birthday. The article is immediately notable in that it is first of seven written by Pauling and his collaborators on the nature of protein and published in that single issue of PNAS.
And as it turns out, these seven articles were revolutionary. While the very act of mailing in all seven at once was audacious, their contents solved riddles that many researchers “believed would not be solved for decades.” Max F. Perutz, a competitor of Pauling’s in the field of biochemistry, read all seven papers in one morning, after which he immediately raced to his lab. Utilizing Pauling’s predictions, he was able to conduct experiments that validated the hypotheses proposed by the papers. He wrote to Pauling that
The fulfillment of this prediction and, finally, the discovery of this reflection in hemoglobin has been the most thrilling discovery of my life.
“The Structure of Proteins: Two Hydrogen-Bonded Helical Configurations of the Polypeptide Chain” was presented as the lead article in the April-May 1951 PNAS. The main question asked by the paper was: What is the structure of polypeptide chains – the backbones of proteins? Pauling, Corey, and Branson claimed that the best way to determine the answer to this question was to acquire an “accurate determination of the crystal structure of amino acids, peptides, and other simple substances related to proteins.” By determining the attributes of these components, which acted as the building blocks of polypeptide chains, the researchers could then make reasonable estimations of what the finished product would look like. Pauling’s group was specifically searching for the interatomic distances between molecules, the bond angles of the chemical bonds, and “other configurational parameters” fundamental to the structures.
Pauling and his colleagues felt that their work answered these questions and that they had determined the parameters satisfactorily. They then used their data to determine that their basic shape was a hydrogen-bonded, helical configuration. They wrote that “An amino acid residue (other than glycine) has no symmetry elements…” (In biochemistry, “residue refers to a specific monomer,” which is “a molecule that may bind chemically to other molecules…”) Because the residues lacked symmetry elements which would force the polymer chain into a symmetrical pattern, “…the only [possible] configurations for a chain…are helical configurations.”
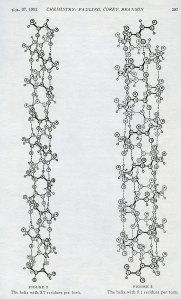
Figures from “The Structure of Proteins: Two Hydrogen-Bonded Helical Configurations of the Polypeptide Chain,” 1951.
Furthermore, the group determined that, based on their calculations of bond angles, there were five possible angles for the helical turns; 165°, 120°, 108°, 97.2°, or 70.1°. Of these, only two, 97.2° and 70.1°, were “reasonable” potential configurations for the polypeptide chain, based on observed interatomic distances. Hydrogen, carbon, oxygen and nitrogen are the elements that make up the polypeptide chain. The chemical bond between C-O and C-N are both double bonds (they have four electrons instead of two). These tight bonds, along with the measured interatomic distances of other components, indicated that the interatomic distances for the chain would have to be small, otherwise the chain would very quickly become unstable. This is the reason that only the 97.2° and 70.1° configurations were acceptable. The other three angles were unstable and would have unraveled because the N-H bonds had too much space between them. Whether the helix turned at a 97.2° or a 70.1° rotation depended on the number of residues per turn in the chain. Pauling and his associates proposed either a 3.7-residue helix or a 5.1-residue helix.
The article ended by explaining why competing hypotheses on the shape of polypeptide chains were incorrect. The article specifically pointed out three hypotheses authored by William Astbury and Florence Bell, William Lawrence Bragg, John Kendrew, and Max Perutz, and Maurice Huggins as being inferior. The Caltech group asserted that each of their models assumed a set number of residues in the polypeptide chains instead of potential variables, and only gave rough estimates of interatomic distances and bond angles. While they all agreed that a helix was the correct shape, the specifics of all other helix models were incorrect because of these deficiencies.
This first paper was just a piece of the larger argument that Pauling was making. Each article was in itself useful, but only when considering the larger sum of the full publication bloc could the full import and implications of Pauling’s work be made visible. Pauling’s thinking proved to be revolutionary and controversial, as such ideas often are. William Lawrence Bragg, a key competitor of Pauling’s, was especially critical. He felt numerous of the Caltech group’s ideas to be outright false, and even the most solid of Pauling’s assertions to be just baby-steps rather than major breakthroughs. Pauling, naturally, disagreed.
