In our new study, we curated > 39,000 amphibian mitochondrial DNA (mtDNA) sequences from GenBank, identified > 2,000 sequencing and taxonomic errors, and published the quality-checked records as a curated dataset with an automated workflow in R. High-quality genetic data should help quantify and protect the diversity of the most threatened vertebrate group on Earth.

Upper left: species of Boophis from Andasibe, Madagascar. Upper right: Dendropsophus anceps from State of Rio de Janeiro, Brazil. Lower left; Dendropsophus bipunctatus from State of Rio de Janeiro, Brazil. Lower right: Bufo bufo from Gelderland, The Netherlands. All images from the author.
Scientists from a broad range of biological disciplines use genetic information like DNA sequences to test ecological and evolutionary hypotheses. Critically, genetics are today essential for naming species and therefore quantifying biodiversity, as well as determining where species live and how many individuals of a species occur in the wild.
Researchers are routinely asked, and more recently frequently required, by scientific journals to submit their DNA sequences to GenBank (among other public repositories of genetic data) as a requirement for publishing a paper. Although GenBank provides some quality controls (e.g., to filter sequences with bacterial contaminants and those from other kingdoms), authors are responsible for the quality of their genetic data and have full freedom to assign these to species in the taxonomy database of GenBank. Notably, once sequences have been deposited in GenBank, records are rarely updated in light of identified errors often resulting from taxonomic progress.
Two important notions emerge from the former status quo:
- The taxonomy of GenBank records has long remained static and therefore records of species known under different names were not inter-linked. However, a recent tool for taxonomic curation (1) still does not update subspecies that are elevated to species.
- GenBank has not put in place a straightforward protocol whereby users can flag errors in deposited sequences on the sequence’s page.
We have just published a study in Nature’s Scientific Data (2) in which we assess the quality of, and curate errors in, GenBank’s records for cytochrome-b gene (cytb) — a gene within the widely studied mitochondrial DNA genome involved in energy transport in eukaryotic cells. We focused on amphibians because these ectotherms are highly diverse and the most threatened vertebrate group with many species facing imminent extinction. Consequently, error-free genetic data are essential for our understanding of their current and future diversity (3).
In our paper, we followed six systematic steps of data-quality assessment and curation:
- we downloaded all GenBank’s Amphibian cytb sequences,
- aligned them against the reference mitochondrial genome of the African clawed frog (an amphibian model organism),
- assessed and corrected (if necessary) the taxonomy of all matches using the Amphibian Species of the World database,
- visually assessed sequences of all species for potential sequencing mistakes or missing nucleotides,
- identified sequences with potential errors using a percentage threshold of DNA differences (~ 3% for amphibians) (4), and
- used the scientific literature to correct faulty taxonomic identity and/or genetic integrity.
Overall, we have curated > 39,000 records and updated the taxonomy of 36,514 cytb sequences in the self-contained dataset ACDC, in honor of the well-known rock band (5). Additionally, we developed an automated workflow in R that (i) retrieves records directly from GenBank using taxonomic key words (which can be tailored to any species or biological group), (ii) removes duplicate records, (iii) creates individual sequence repositories per species, and (iv) flags sequences that could include errors. Both the database and the R workflow are freely available in figshare (see here).
We classified a total of 2,359 incorrect sequences (6%) into specific categories depending on the nature of the error. Those categories comprised incorrect taxonomy, hybridised specimens, genetic contamination, chimeras, sequencing errors, and submission errors. Taxonomic misidentifications outnumbered other error categories (84%), some occurring before and others after data submission to GenBank as taxonomy progresses. Overall, we updated the taxonomy of > 4,800 sequences (13% of the raw dataset).
We recommend the following to improve data quality in GenBank:
- GenBank could create a friendly online tool, accessible from each record’s page, through which the community (including GenBank staff and sequence owners) could communicate errors affecting individual records.
- Authors could match the taxonomic identity in each GenBank record versus the publication when first reporting the sequence, and mismatches could be flagged in GenBank.
- Authors could, prior to submission, align their sequences against GenBank records using the online BLAST tool, and visually assess mutation patterns to curate the taxonomy and sequence quality of their submissions.
- Authors should not only cite GenBank accession numbers in the publications using the data for the first time, but therein also report (i) full taxonomic identity (family, genus, species, subspecies), (ii) study locality and geo-position of samples, (iii) phylogenetic group/clade/lineage, and (iv) a legend linking samples to manuscript figures and/or tables. And scientific journals could request the above information as a compulsory requirement for publication.
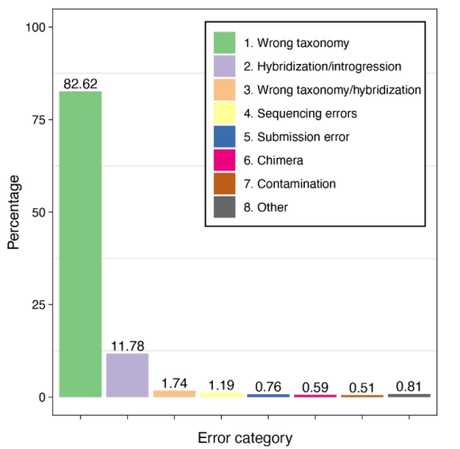
Percentage distribution of error categories for 2,359 GenBank Amphibian cytochrome b sequences identified as erroneous (2). ‘Wrong taxonomy’ = misidentification or updated taxonomy. ‘Hybridization/Introgression’ = taxonomic misidentification due to those processes along with incomplete lineage sorting. ‘Sequencing errors’ = ambiguous nucleotides. ‘Submission error’ = sequence or metadata inconsistent with original manuscript. ‘Chimera’ = multiple sets of DNA resulting from artefacts. ‘Contamination’ = non-amphibian (partial) sequences. ‘Others’ = mostly high intraspecific divergence suggesting sequencing errors.
The scientific community has been long aware of prevailing errors in GenBank (6), but few authors have quantified the magnitude of this problem (e.g., 7, 8). Erroneous genetic records can hamper the correct identification of species and affect scientifically informed conservation and management actions.
Consequently, our findings are strongly relevant to amphibians (2) because this group faces a range of threats; particularly: (i) global spread of deadly fungal diseases (9, 10), (ii) rapidly changing environmental temperatures and water availability (11), and (iii) destruction of habitat (12).
Critically, some species have become extinct even before they were described (13), which could especially threaten tropical species (14, 15). We hope to stimulate better data-handling practices in the scientific community working on these spectacular animals.
Matthijs P. van den Burg / [email protected]
References
- Schoch, CL et al. (2020). NCBI Taxonomy: a comprehensive update on curation, resources and tools. Database 2020: baaa062
- van den Burg, MP et al. (2020). ACDC, a global database of amphibian cytochrome-b sequences using reproducible curation for GenBank records. Data 7: 268
- Vences, M et al. (2005). Deciphering amphibian diversity through DNA barcoding: chances and challenges. Trans. R. Soc. Lond. B Biol. Sci. 360: 1859-1868
- Köhler, J et al. (2005). New amphibians and global conservation: a boost in species discoveries in a highly endangered vertebrate group. BioScience 55: 693-696
- van den Burg, MP et al. (2020). ACDC, a global database of amphibian cytochrome-b sequences using reproducible curation for GenBank records. https://doi.org/10.6084/m9.figshare.9944759.v4
- Harris, D. (2003). Can you bank on GenBank? Trends Ecol. Evol. 18: 317-319
- Li, X et al. (2018). Detection of potential problematic cytb gene sequences of fishes in GenBank. Front. Genet. 9: 30
- Layer, M et al. (2019). GenBank is a reliable resource for 21st century biodiversity research. Proc. Natl. Acad. Sci. USA 116: 22641-22656
- Martel, A et al. (2014). Recent introduction of a chytrid fungus endangers Western Palearctic salamanders. Science 346: 630-631
- Crawford, AJ et al. (2010). Epidemic disease decimates amphibian abundance, species diversity, and evolutionary history in the highlands of central Panama. Proc. Natl Acad. Sci. USA 107: 13777-13782
- Lips, KR et al. (2008). Riding the wave: reconciling the roles of disease and climate change in amphibian declines. PLoS Biol. 6: e72
- Cushman, SA. (2006). Effects of habitat loss and fragmentation on amphibians: a review and prospectus. Biol. Conserv. 128: 231-240
- Jaramillo, AF et al. (2020). Vastly underestimated species richness of Amazonian salamanders (Plethodontidae: Bolitoglossa) and implications about plethodontid diversification. Mol. Phylogenet. Evol. 149: 106841
- Vieites, DR et al. (2009). Vast underestimation of Madagascar’s biodiversity evidenced by an integrative amphibian inventory. Proc. Nat. Acad. Sci. USA 106: 8267-8272
- Rosa, IMD et al. (2016). The environmental legacy of modern tropical deforestation. Curr. Biol. 26: 2161-2166

