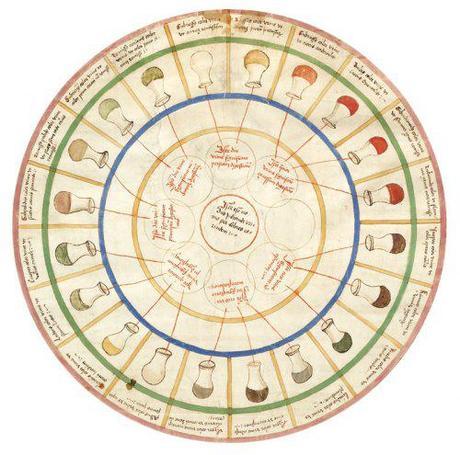
“This urine wheel was published in 1506 by Ullrich Pinder, in his book Epiphanie Medicorum. It describes the possible colours, smells and tastes of urine, and uses them to diagnose disease.”
Jeremy K. Nicholson & John C. Lindon Nature 2008 (1)
Metabolomics has come a long way since the days of tasting urine. Now techniques like high-throughput mass spectrometry can provide us detailed information about the small molecules we find in the body–which can be influenced by diet, environmental exposure (to toxins, medicine, etc), or even the gut microbiome. Metabolomics will also capture the genetic or epigenetic differences in how individuals process these exposures.
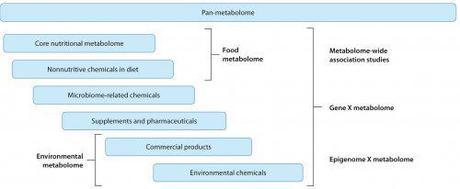
Dean P. Jones, et al. Annual Reviews (2012)
Metabolome-Wide Association Studies
New high-throughput technologies have great potentials is unbiased case-control studies looking at as many metabolites as possible–Metabolome Wide Association Studies (similar to the GWAS studies done in genetics). These studies would look for the association of any metabolite with a given disease–after which research into that metabolite could indicate a variety of causes–nutritional differences, environmental exposure, or even mutations in metabolic pathways.
Any given study would not be the end all, as metabolomic exposure occurs over a life-time. Other important factors may include exposures while in the womb to teratogens–substances that interfere with the development of an embryo or fetus that cause birth defects.

The most famous example of a teratogen is the drug Thalidomide
Metabolomics and Neursocience
Research into metabolomics is in it’s infancy and not much research has been so far relating it to psychiatric or neurodegenerative disease. The GWAS for psychiatric disease were not as fruitful as was hoped–probably due to both the complex natures of the diseases and also the murkiness of classification associated with psychiatric disease. The same classification problems will make MWAS difficult, but could help explain the enviornmental factors in those diseases (e.g. why one genetically identical twin gets schizophrenia while another doesn’t).
MWAS may also identify biomarkers which can be used in either the identification of psychiatric disease, or subcategorization of a disease. This would allow more reproducible, systematic studies of the disease and help seperate similarly-manifesting diseases by their root cause.
We really have a wealth of riches in terms of high-data yielding approaches to tackle these diseases: genomics, epigenomics, metabolomics, and brain-imaging, but unfortunately these methods remain expensive and time consuming. Currently one of the biggest hold-ups in metabolomics is man-power on the bioinformatics side of interpretting the mass/spec results. There are hundreds of thousands of metabolites in what we eat alone and we are only scratching the surface.
References
1. Nicholson, J. K. and J. C. Lindon (2008). “Systems biology: Metabonomics.” Nature 455(7216): 1054-1056. http://dx.doi.org/10.1038/4551054a
2. Jones, D. P., Y. Park, et al. “Nutritional Metabolomics: Progress in Addressing Complexity in Diet and Health.” Annual Review of Nutrition 32(1): 183-202. http://www.annualreviews.org/doi/full/10.1146/annurev-nutr-072610-145159

