This study has been popping up all over the AMR radar for me for the past week; so, when I was stuck in traffic today, I decided to sit down and give the article a read. An interesting read, which does provide insights into the rapid global spread of AMR, this article, however, has been taken way out of context by the headlines of tabloids screaming the end-of-the-world prophecies. Now, I am the last person to discount the threat of AMR, but at the same time, perhaps this is not as bad as the tabloids made it out to be. The worrisome thing remains that we are no strangers to the introduction of “invasive” pathogens in previously naïve settings, which end up wreaking havoc over the years. Perhaps the cholera experience of Haiti stands out to be one of the most remarkable examples of this mishap. Finding antimicrobial resistance genes (ARGs) and mobile genetic elements (MGEs) which are definitely not of local origin does raise the specter of an invasive, allochthonous route, but before hitting the panic bells, we need more evidence on the routes and sources for the emergence of these unlikely tourists in the wintry north.
In the study, the authors looked at forty samples from across 8 soil clusters, as highlighted in the image below.
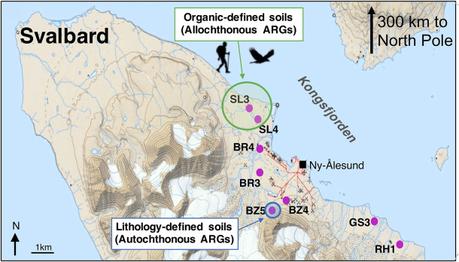 Sampling locations of the eight soil clusters in Kongsfjorden. Topographic map has been adapted from the Norwegian Polar Institute.
Sampling locations of the eight soil clusters in Kongsfjorden. Topographic map has been adapted from the Norwegian Polar Institute.Barely a stone’s throw away from the North Pole, the study area provided a unique geoclimatic ecosystem in which to study the presence of AMR genes in the soil. Located on a remote island, with no native agriculture or industry, the small resident population (n=120) of Kingsfjorden, Svalbard, live in an area which is cold enough to preserve DNA in soil for a prolonged period, yet, never freezes over due to Gulf currents. The area is, thus, exposed to importation of ARGs and MGEs, piggybacking on migratory birds or travelers who may pass through this Arctic sentinel.
The authors report that they identified 131 ARGs in the forty areas studied, and of these, they found 39 in all the clusters – these likeluy represented the autochthonous ARGs which were likely background presence, irrespective of anthropogenic threats. The study does not clearly define a strategy to understand and define whether an ARG is autochthonous or allochthonous. The definition used by the authors to designate particular ARGs as allochthonous include the fact that the ones so labelled were isolated from only a few of the sampled clusters and were associated with resistance against antibiotics which are of critical importance for human health (aminoglycosides, macrolides, carbapenems, penicillin, and cephalosporins). This definition, whereby the putative allochthonous versus autochthonous status is ascertained based on the presence or absence of each ARG compared across the sampled clusters, could potentially be rife with controversies. Although such a definition allowed the authors to pin point the presence of “foreign” ARGs and MGEs in the Arctic environment, it will also feed into the uncertainties about the sources of the same.
Resistance genes were detected against nine major classes of antibiotics, though the predominating ones were against aminoglycosides and beta-lactams. In five out of the eight soil clusters sampled, the researchers detected the blaNDM-1 gene. This gene, which produces the New Delhi metallo-β-lactamase (NDM-1) enzyme, which confers resistance against carbapenems, is definitely not a local entity and has likely traveled over on biological vehicles. Isolation of blaNDM-1 virtually seals the case for the international migration of AMR. This report is also the first time that this resistance gene has been demonstrated in the High Arctic areas. More concerning was the fact that in one of the locations, SL3, shown in green circle above, the authors discovered a painful secret:
Levels of both blaNDM-1 and FabK were approximately 100× greater in
SL3 than in the other clusters. Indeed, relative abundances of blaNDM-1
at SL3 were similar to levels found in environments impacted by hospital
and urban wastewaters.
This, in combination with the fact that the authors found an “AMR gradient” across the study site, points to issues which need to be studied in more details. Despite the absence of any local, AMR producing drivers, the fact that there is differential occurrence of ARGs and MGEs in the soil, indicates some differential levels of “AR pollution” in the area, according to the authors. I wholeheartedly agree with this contention of the authors, and in fact, find it more troubling than the mere presence of the blaNDM-1 genes in the area. The presence of “Indian origin” AMR genes in the Arctic makes for sensational headlines and clickbait traps, but far greater is the implication hidden in the small sentence where the authors ponder on why the differential ARG gradient was observed in an area where the traditional drivers and determinants of AMR are not at play.
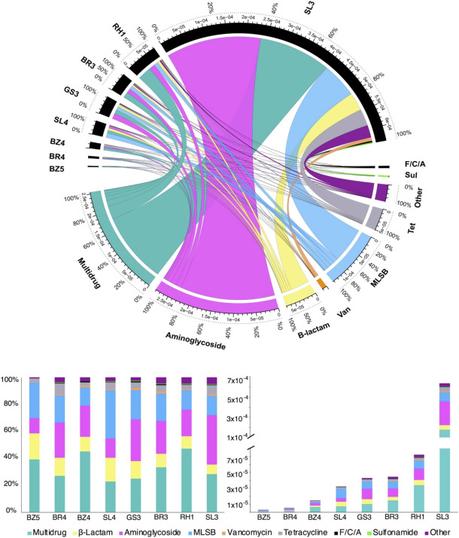 Distribution and abundance of ARGs in the eight High Arctic clusters classified by antibiotic class and cluster location. The Chord diagram presents the abundance of ARGs in each sample that associate with each respective antibiotic class among the eight soil clusters (top). Presented below the diagram is the relative abundance of ARGs normalized to the total abundance of 16S rRNA gene (bottom right) and relative proportions of ARG classes in each cluster (bottom left). MLSB denotes Macrolide-Lincosamide-Streptogramin B. Other labels represent ARGs that do not have a direct antibiotic class. F/C/A denotes florcholoram-phenicol antibiotic class.
Distribution and abundance of ARGs in the eight High Arctic clusters classified by antibiotic class and cluster location. The Chord diagram presents the abundance of ARGs in each sample that associate with each respective antibiotic class among the eight soil clusters (top). Presented below the diagram is the relative abundance of ARGs normalized to the total abundance of 16S rRNA gene (bottom right) and relative proportions of ARG classes in each cluster (bottom left). MLSB denotes Macrolide-Lincosamide-Streptogramin B. Other labels represent ARGs that do not have a direct antibiotic class. F/C/A denotes florcholoram-phenicol antibiotic class.The authors have adequately focused on the opportunity presented by these scantily populated outer fringes of human civilization as sentinels of globalization of AMR; however, without a better understanding of how the ARGs got there within a mere 2-3 years of their identification, such a sentinel surveillance will provide minimal actionable evidence.
This study, which has captured the attention of the popular media, points to a deeper malady which is getting buried under all the noise around blaNDM-1 and its geopolitical connotations. One can only hope that the scientific enquiries, which should be precipitated by this publication, are not derailed or disorientated by the surrounding brouhaha.
